Math Genealogy Project
I traced my mathematical lineage back into the XIV century at The Mathematics Genealogy Project. Imagine my surprise when I discovered that a big branch in the tree of my scientific ancestors is composed not only by mathematicians but also by big names in the fields of Physics, Chemistry, Physiology and even Anatomy.
There is some blue blood in my family: Garrett Birkhoff, William Burnside (both algebraists). Archibald Hill, who shared the 1922 Nobel Prize in Medicine for his elucidation of the production of mechanical work in muscles. He is regarded, along with Hermann Helmholtz, as one of the founders of Biophysics.
Thomas Huxley (a.k.a. “Darwin’s Bulldog”, biologist and paleontologist) participated in a famous debate in 1860 with Samuel Wilberforce, Lord Bishop of Oxford. This was a key moment in the wider acceptance of Charles Darwin’s Theory of Evolution.
There are some hard-core scientists in the XVIII century, like Joseph Barth and Georg Beer (the latter is notable for inventing the flap operation for cataracts, known today as Beer’s operation).
My namesake Franciscus Sylvius, another professor in Medicine, discovered the cleft in the brain now known as Sylvius’ fissure (circa 1637). One of his advisors, Jan Baptist van Helmont, is the founder of Pneumatic Chemistry and disciple of Paracelsus, the father of Toxicology (for some reason, the Mathematics Genealogy Project does not list any of these two in my lineage—I wonder why).
There are other big names among the branches of my scientific genealogy tree, but I will postpone this discovery towards the end of the post, for a nice punchline.
Posters with your genealogy are available for purchase from the pages of the Mathematics Genealogy Project, but they are not very flexible neither in terms of layout nor design in general. A great option is, of course, doing it yourself. With the aid of python, GraphViz and the sage library networkx, this becomes a straightforward task. Let me show you a naïve way to accomplish it:
Let us start by searching for a name in the database online. Once in screen, note the string of numbers at the end of the URL obtained:
http://genealogy.math.ndsu.nodak.edu/id.php?id=113998Every individual in the database has a unique ID in this fashion. Note also, in the source of the page, the field Advisor: If the advisor of an individual is not Unknown, the page will link to the corresponding page. We can easily retrieve the ID of the advisor from the source code. Even better, we can code a small script in python to recursively go upwards in the database gathering the ID’s of your ancestors in a dictionary. One fast way to accomplish this could be as follows:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
import urllib
def augment_genealogy(subject_id,subject_tree):
# This function assumes that subject_id in not in subject_tree
f = urllib.urlopen("http://genealogy.math.ndsu.nodak.edu/id.php?id="+subject_id)
subject = f.read()
f.close()
if not subject.count("Advisor: Unknown"):
# How many advisors did subject have?
# For each advisor, retrieve their information, and attach
# them to subject as parents
advisor_list = []
for advisor in range(subject.count("Advisor")):
subject = subject.partition("Advisor")[2].partition("id=")[2]
advisor_id = subject[0:subject.index("\"")]
advisor_list.append(advisor_id)
if advisor_id not in subject_tree:
augment_genealogy(advisor_id, subject_tree)
subject_tree[subject_id] = advisor_list
return subject_tree
else:
subject_tree[subject_id]=[]
return subject_tree
But what good is a genealogy tree if we cannot see the names of the ancestors? The simple script below takes care of retrieving the names online for each of the IDs in our previously created dictionary. Of course, one could instead include the appropriate string manipulation in the retrieval script above, and kill two birds with one stone.
1
2
3
4
5
6
7
8
# Create a dictionary that maps to each id, its name
names = dict()
for id in data:
f = urllib.urlopen("http://genealogy.math.ndsu.nodak.edu/id.php?id="+id)
s = f.read()
f.close()
name = s.partition("The Mathematics Genealogy Project - ")[2]
names[id] = unicode(name[0:name.index(">")].strip(), "ascii", "ignore").encode()
These are the names in my tree, for example: Do you recognize any of them?
>>> names.values()
['Adam Sedgwick', 'Adolph Vorstius', 'Adriaan van den Spieghel', 'Alessandro
Sermoneta', 'Alexander Hegius', 'Andreas Vesalius', 'Angelo Poliziano', 'Anton
von Strck', 'Antonio Brasavola', 'Antonius Thysius', 'Archibald Hill', 'Arnold
Senguerdius', 'Basilios Bessarion', 'Benjamin Pulleyn', 'Bradley Lucier',
'Burchard de Volder', 'Cristoforo Landino', 'Daniel Berckringer', 'Demetrios
Chalcocondyles', 'Demetrios Kydones', 'Desiderius Erasmus', 'Duncan Liddel',
'Edward Waring', 'Elissaeus Judaeus', 'Emmanuel Stupanus', 'Erasmus Reinhold',
'Francisco Blanco-Silva', 'Franciscus Gomarus', 'Franciscus Sylvius', 'Franck
Burgersdijk', 'Franois Du Jon, Sr.', 'Franois Dubois', 'Gabriele Falloppio',
'Gaetano da Thiene', "Galileo de' Galilei", 'Garrett Birkhoff', 'Gemma
Frisius', 'Georg Beer', 'Georgios Gemistos', 'Georgius Hermonymus', 'Gerard van
Swieten', 'Gilbert Jacchaeus', 'Gilles de Roberval', 'Girolamo Aleandro',
'Gisbertus Voetius', 'Guarino da Verona', 'Guillaume Bud', 'Henricus Renerius',
'Henry Bracken', 'Herman Boerhaave', 'Hieronymus Fabricius', 'Immanuel
Tremellius', 'Isaac Barrow', 'Isaac Newton', 'Jacob ben Jehiel Loans', 'Jacobus
Arminius', 'Jacobus Latomus', 'Jacobus Sylvius', "Jacques Lefvre d'taples",
'Jacques Toussain', 'Jakob Milich', 'Jan Standonck', 'Jan van Campen', 'Janus
Lascaris', 'Jean Tagault', 'Jerry Bona', 'Johann Reuchlin', 'Johannes
Argyropoulos', 'Johannes Hommel', 'Johannes Polyander van Kerckhoven',
'Johannes Stffler', 'Johannes Sturmius', 'Johannes de Bruyn', 'Johannes von
Andernach', 'John Craig', 'John Cranke', 'John Dawson', 'John Langley', 'Joseph
Barth', 'Karl Pearson', 'Leo Outers', 'Maarten van Dorp', 'Manuel Chrysoloras',
'Marsilio Ficino', 'Martinus Schoock', 'Matteo Realdo Colombo', 'Matthaeus
Adrianus', 'Melchior Wolmar', 'Micheal Foster', 'Moses Perez', 'Niccol
Leoniceno', "Niccolo' Fontana", 'Nicolas Clnard', 'Nilos Kabasilas', 'Ognibene
Bonisoli da Lonigo', 'Ostilio Ricci', 'Paul Wittich', 'Pelope', 'Petrus
Curtius', 'Petrus Ramus', 'Petrus Ryff', 'Philip Hall', 'Philipp Melanchthon',
'Pietro Roccabonella', 'Ralph Fowler', 'Robert Smith', 'Roger Cotes', 'Rudolf
Agricola', 'Rudolph Snellius', 'Rutger Rescius', 'Scipione Fortiguerra', 'Sir
Francis Galton', 'Stephen Whisson', 'Theodor Zwinger', 'Theodoros Gazes',
'Theodorus Beza', 'Thomas Kempis', 'Thomas Huxley', 'Thomas Jones', 'Thomas
Jones', 'Thomas Postlethwaite', 'Ulrich Zasius', 'Valentin Thau', 'Valentine
Naibod', 'Vincenzio Viviani', 'Vittorino da Feltre', 'Walter Fletcher', 'Walter
Taylor', 'William Hopkins', 'William McKenzie', 'William Sharpey', 'Wolferd
Senguerdius']Let us move to sage and plot a graph of what we have obtained. Let us try with my ID ("113998"). We have different graphing options with networkx:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
import networkx
import matplotlib.pyplot
data = augment_genealogy("113998",dict())
Tree = networkx.MultiDiGraph(data)
# First graph
matplotlib.pyplot.figure(figsize=(50,50))
pos = networkx.spring_layout(Tree, iterations=900)
networkx.draw(Tree, pos, node_size=0, alpha=0.4, font_size=24)
matplotlib.pyplot.savefig('/Users/blanco/Desktop/tree.png')
# Second graph
matplotlib.pyplot.figure(figsize=(50,50))
pos = networkx.shell_layout(Tree)
networkx.draw(Tree, pos, node_size=0, alpha=0.4, font_size=24)
matplotlib.pyplot.savefig('/Users/blanco/Desktop/tree.png')
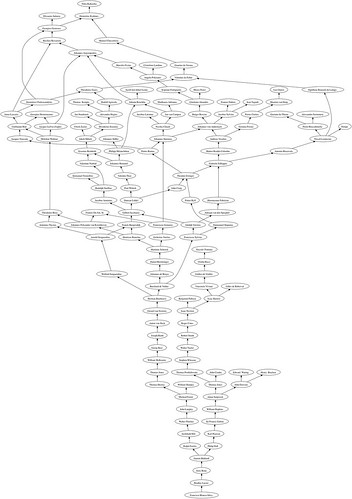
Because of the cooperative nature among scientists, it is not unusual to encounter several ancestors over different generations linked to the same advisor. This usually destroys the structure of binary tree that one would expect for this type of graph, making the plotting of the data quite a challenge.
T = Graph(data)
T.is_tree()FalseAn easy workaround is to install GraphViz, and use interaction to this software from the networkx libraries. The following script attaches to each label the corresponding name, and produces the desired genealogy tree:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
T = DiGraph(data)
T.graphviz_to_file_named('/Users/blanco/Desktop/tree.dot')
# Here is where we change the labels from numbers to names
# We need to do it this way to avoid the issue "different people with same name"
fr = open("/Users/blanco/Desktop/tree.dot", "r")
workingString = fr.read()
fr.close()
newString = str()
while workingString.count("[label=\""):
breakString = workingString.partition("[label=\"")
newString += breakString[0] + breakString[1]
stepString = breakString[2]
newString += names[stepString[0:stepString.index("\"")]]+"\""
workingString = stepString.partition("\"")[2]
newString += workingString
fw = open("/Users/blanco/Desktop/newtree.dot", "w")
fw.write(newString)
fw.close()
os.system('open -a /Applications/GraphViz.app /Users/blanco/Desktop/newtree.dot')