Today I would like to show a very basic technique of detection based on simple convolution of an image with small kernels (masks). The purpose of these kernels is to enhance certain properties of the image at each pixel. What properties? Those that define what means to be an edge, in a differential calculus way—exactly as it was defined in the description of the Canny edge detector. The big idea is to assign to each pixel a numerical value that expresses its strength as an edge: positive if we suspect that such structure is present at that location, negative if not, and zero if the image is locally flat around that point. Masks can be designed so that they mimic the effect of differential operators, but these can be terribly complicated and give rise to large matrices. The first approaches were performed with simple \( 3 \times 3 \) kernels. For example, Faler came up with the following four simple masks that emulate differentiation:
Note that, adding all the values of each matrix, one obtains zero. This is consistent with the third property required for our kernels: in the event of a locally flat area around a given pixel, convolution with any of these will offer a value of zero.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
from numpy import *
from scipy import misc, ndimage
def detect_edges(image,masks):
edges=zeros(image.shape)
for mask in masks:
edges=maximum(scipy.ndimage.convolve(image,mask), edges)
return edges
image=scipy.misc.lena()
Faler=[ [[-1,0,1],[-1,0,1],[-1,0,1]],
[[1,1,1],[0,0,0],[-1,-1,-1]],
[[-1,-1,-1],[-1,8,-1],[-1,-1,-1]],
[[0,1,0],[-1,0,1],[0,-1,0]] ]
edges=detect_edges(image, Faler)
 |
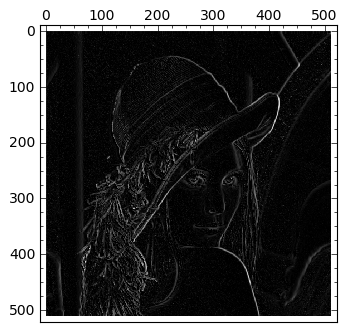 |
Note that the computed edges show different intensities; the brighter, the more likely to be a genuine edge—as found by this technique. With a simple thresholding we should be able to create a True-False image with the required edge map. How would the reader proceed to find a reliable threshold for this task?
Sobel pushed the technique a little bit further by creating compass gradients. In a nutshell: at any given pixel, an edge could spread in eight different directions; therefore, we could create for each possibility a different kernel capturing such event. These are the masks proposed by Sobel, following that philosophy:
You have surely heard also of Kirsch and Prewitt’s edge detectors. They can also be realized as convolutions with the proper sets of kernels in the same way. I collected below their corresponding masks for reference. I leave to the reader the task to run a few experiments, compare the different outputs for the same image, and find good choices of thresholds for each case. I also encourage you to create your own set of masks: do not restrict yourself to \( 3 \times 3 \) matrices, or only to eight possibilities. Do you see any connection with wavelets? If so, how would you combine these two a-priori disconnected ideas to identify edges?
Kirsch = [ [[5,5,5],[-3,0,-3],[-3,-3,-3]],
[[-3,5,5],[-3,0,5],[-3,-3,-3]],
[[-3,-3,5],[-3,0,5],[-3,-3,5]],
[[-3,-3,-3],[-3,0,5],[-3,5,5]],
[[-3,-3,-3],[-3,0,-3],[5,5,5]],
[[-3,-3,-3],[5,0,-3],[5,5,-3]],
[[5,-3,-3],[5,0,-3],[5,-3,-3]],
[[5,5,-3],[5,0,-3],[-3,-3,-3]] ]
Prewitt = [ [[1,1,1],[1,-2,1],[-1,-1,-1]],
[[1,1,1],[1,-2,-1],[1,-1,-1]],
[[1,1,-1],[1,-2,-1],[1,1,-1]],
[[1,-1,-1],[1,-2,-1],[1,1,1]],
[[-1,-1,-1],[1,-2,1],[1,1,1]],
[[-1,-1,1],[1,-2,1],[1,1,1]],
[[-1,1,1],[-1,-2,1],[-1,1,1]],
[[1,1,1],[-1,-2,1],[-1,-1,1]] ]